Thanks for your comments, I’ve noted some iumporvements to documentation under SUP-789 and whilst speaking with one of the Developers he pointed me to this reference.
https://cran.r-project.org/web/packages/nmw/nmw.pdf
(Understanding Nonlinear Mixed Effects Modeling for Population Pharmacokinetics)
R Matrix R matrix of NONMEM, the second derivative of log likelihood function with respect to estimation parameters
The R matrix is the Fisher information matrix constructed from the second derivative of the objective function with respect to the various parameters estimated.
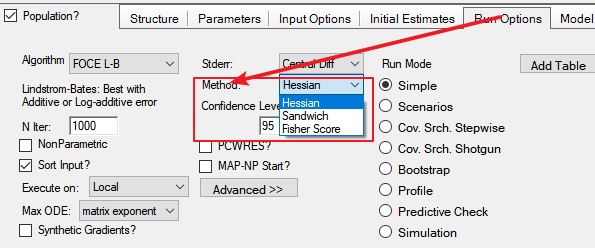
R matrix is the same as Hessian in NLME
S Matrix S matrix of NONMEM, sum of individual cross-product of the first derivative of log likelihood function with respect to estimation parameters
The more approximate S matrix (score matrix) is constructed from the first derivatives of each individual’s objective function contribution with respect to the various parameters estimated
S matrix is the same as Fisher score in NLME
The variance–covariance of the estimates is evaluated as R−1 or S−1. By default the variance–covariance is evaluated as R-1SR-1 (Sandwich in NLME**)**
Note that in 8.2 new option ‘AutoDetect’ is added.
When selected, NLME automatically chooses the standard error calculation method. Specifically, if both Hessian and Fisher score methods are successful, then it uses the Sandwich method. Otherwise, it uses either the Hessian method or the Fisher score method, depending on which method is successful. The user can check the Core Status outputs to see which method is used.
Simon.
PS for the ‘nerds’ ;0), you can review internals of calculations using debug CovStep() function of that package.