Hi, Can someone help me out on the modeling for the following rat oral data? The data suggested a 2 compartment model. I tried running the WNL classical PK model: Extravascular 1st order 2 compartment w lag time but failed in all attempts. Timepoints (hr): 0, 1.667, 0.333, 0.5, 1, 2, 4, 6, 24 Concentration (ng/mL) 0, 11874.803, 10244.172, 6821.271, 2464.625, 495.161, 145.776. 93.997, 2.789 Dose = 75 mg Thanks John [file name=data.xlsx size=8456]Certara | Drug Development Solutions (8.26 KB)
Dear John I think that it is important to use one of my cook books to fit models to data with minimum risk of crashs.. Here is the deal. You always start with NCA. use Ka=10lambdaz and use V and Cl obtained from NCA for the next step. step 2: 1 compt model, clearance parametrizarion,use multiplicative error, 0.1 for stdev, Cl and V from NCA and Ka=lambdaz10 step3: add mdv column as at t=0, it is a nuisance response of 0 which is not necessary to fit and exact 0 values for responses are always problem step4: after you fit with the 1 c, copy the model to the workflow and accept all fixed and random Then shit to 2c with V2=V and Cl2=Cl fit that model to the dataset: step5:add tlag with initial estimate around the first time point at which the observation s not zero add a table to nicely comparing observed versus predicted I did all that and it looks great to me. let me know if you need further help. The project is attached. Best Serge [file name=john.phxproj size=1383067]Certara | Drug Development Solutions (1.32 MB)
Thank you Sergeguzy. I am still struggling with the new Phoenix Modelling feature so it takes a while for me to get used to it. Back to the data, 2 questions. 1) So just take V and Cl (V1, Cl1, V2, Cl2) to compute the 2 Kels? 2) You mentioned in your description about 0 values for response. What would you do to concentration at the end of the profile (example, 24hr) that is BLQ? I usually set it as 0. Thanks John
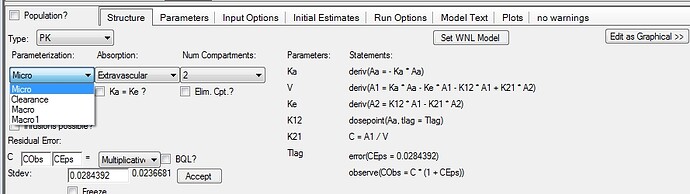
John, Serge built the models parametised as Clearance as that seems to be the way that most groups prefer but alternatively you could have built it as a micro constant model;  Either way you can actually define other desired output as Secondary parameters and, IF you are using models that were inthe WNL classic library then the Phoenix model object can actually write them for you; see the use of “set WNL model” button;
Either way you can actually define other desired output as Secondary parameters and, IF you are using models that were inthe WNL classic library then the Phoenix model object can actually write them for you; see the use of “set WNL model” button;  Lastly I would NOT set a BQL as 0, if you leave as BQL or other character it is simply treated as missing, or if you prepare your data a little more you can actually incorporate it into your model.
Lastly I would NOT set a BQL as 0, if you leave as BQL or other character it is simply treated as missing, or if you prepare your data a little more you can actually incorporate it into your model.  (there are some discussions on using this already on here) Simon
(there are some discussions on using this already on here) Simon 
Dear John I added the “set Winnonlin models” as suggested by Simon and played a little bit. Look at the last model. In the results, look at the secondary parameters and you will get ke, k12 and k21 as you wanted. note that I had to delete some model parameters computations because they use different setting than the new phoenix Winnonlin. Let me know if you need further clarifications. best Serge [file name=john-20140902.phxproj size=1716042]Certara | Drug Development Solutions (1.64 MB)
Thank you guys, I toyed around the program over the weekend with the Set WNL option and figured out the 2ndary parameters. One more question. How does the program handle the datapoint if I chose MDV? Example, as Simon has said, don’t put 0 for BQL data. What about the BQL at time 0? If I am certain that there is no drug at time 0 then should I set time 0 as 0? Should I tag time 0 concentration (which is 0) as MDV? Thanks John
One more question. How does the program handle the datapoint if I chose MDV? Look at my data set with mdv. mdv=1 means that the program will not use that observation. mdv=0 means that the observation is used. In addition you use input options and click on mdv checkbox (look at it). You never put bql if the true value is 0 as bql means that the value is between 0 and LOQ. Example, as Simon has said, don’t put 0 for BQL data. What about the BQL at time 0? If I am certain that there is no drug at time 0 then should I set time 0 as 0? No, just put mdv=1 Should I tag time 0 concentration (which is 0) as MDV? at t=0, you put 1 in the mdv column (look at my data set). Best Serge
Hi Serge, I am still having some issues. Can you try out the attached dataset for me? After completing the modeling (2 compartment, used MDV), I tried to generate a raw data vs fitted concentration table but I could not get an output from 0 - 24 hrs. Thanks again. John [file name=4R3.xls size=10752]Certara | Drug Development Solutions (10.5 KB)